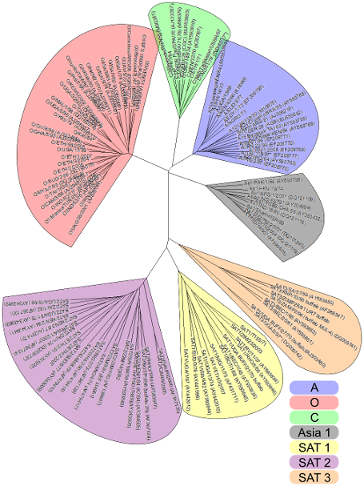
There are 7 different varieties (serotypes) of FMDV, named: Serotypes O, A, C, O, Asia 1, SAT 1, SAT 2 and SAT 3. All give rise to a similar disease and infection but one serotype does not confer immunity against another.
The first three serotypes
In 1922 Vallée and Carré demonstrated that there were two serotypes of FMDV (Vallée & Carré, 1922). They named these Vallée O and Vallée A, after the regions they orginated from (department of Oise in France and Germany [Allemagne in French]).
Four years later Waldmann and Trautwein demonstrated the existance of three serotypes (Waldmann & Trautwein, 1926), which they named Waldmann A, B and C. Waldmann A and Waldmann B were found to be the same as Vallée O and Vallée A, respectively. While Waldmann C remained as a distinct third serotype.
The names of Vallée O, Vallée A and Waldmann C were later shortened to O, A and C.
The next three serotypes

Two new serotypes were identified in 1948 in samples from Bechuanaland (Botswana) and Northern Rhodesia (Zambia) (Brooksby, 1958). Retrospective testing of samples from Southern Rhodesia (Zimbabwe) from the 1930s found both of these new serotypes and a third new serotype as well.
These new serotypes were named Southern African Territories 1 to 3 (abbreviated to SAT 1, SAT 2 and SAT 3).
The final serotype
The serotype Asia 1 was identified in the early 1950s in viruses isolated from India and Pakistan (Dhanda et al., 1957 and Brooksby & Rogers, 1957).
The WRLFMD uses a standard colour scheme for serotypes:
O: Red
A: Blue
C: Green
Asia-1: Grey
SAT 1: Yellow
SAT 2: Purple
SAT 3: Orange
Genetics of the serotypes
Each serotype can be genetically split into various topotypes and lineages.Topotypes and lineages
The WRLFMD suggests the following format for writing the serotype, topotype, and lineage:
SEROTYPE /TOPOTYPE ABBREVIATION/LineageSUB-LINEAGE
(for example: O/ME-SA/PanAsia-2TER-08, A/ASIA/Iran-05HER-10, Asia 1/ASIA/Sindh-08, etc.)
Genetic relationship (using VP1 sequences) between FMDV serotypes. The prototype strain sequences used to create this phylogenetic tree are available on this site
(click on the picture to open a large version in a new window).