Inside the capsid is a 8.4-kilobase, positive-sense, single-stranded RNA genome (making FMDV a Group IV virus in the Baltimore classification) that is covalently bound at its 5′ end to the small viral protein 3B and is polyadenylated at its 3′ end. Upon virus entry into a cell, the viral genome is translated into a polyprotein which is co- and post-translationally cleaved by viral proteinases into 12 mature proteins.
Genome of strain BAN/NA/Ha-156/2013 (serotype O, from Bangladesh)

The FMDV genome organisation is similar to that of other picornaviruses, including a large single open reading frame (ORF) flanked by highly structured 5′ untranslated region (5′ UTR), containing an Internal Ribosome Entry Site (IRES) essential for the translation of the ORF, and a shorter 3' untranslated region (3′ UTR).
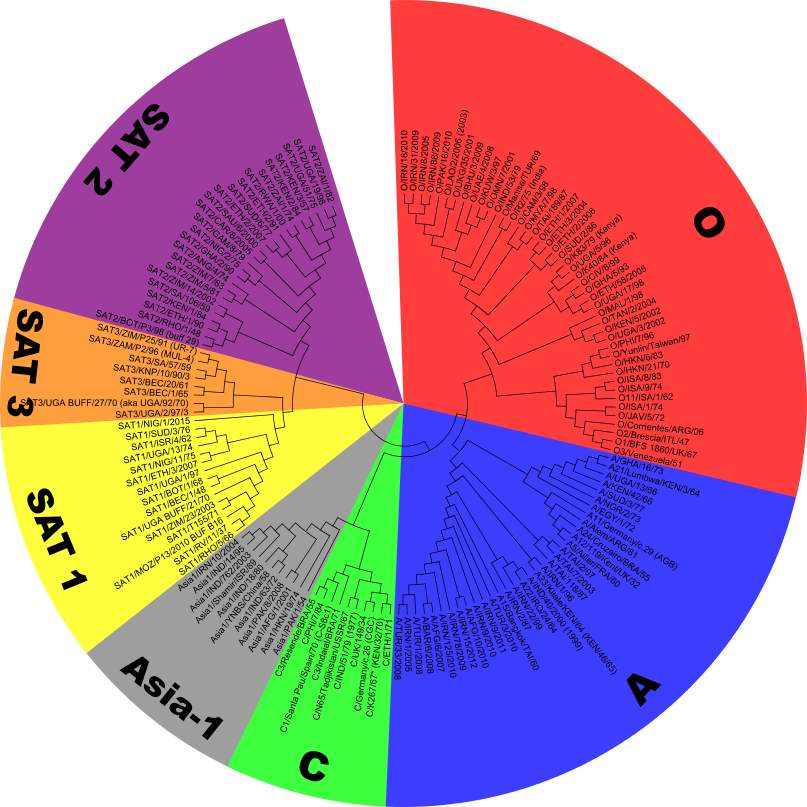
Sequences are available for a large number of strains and sequencing these strains shows the relationship between strains and serotypes.
These sequences can be used to generate a phylogentic tree showing the relationship between the different strains of FMD (see left).